Efficient Python for Data Scientists #3/14: Optimizing Your Numpy & Pandas Code
Learn how to optimize your python code and make it efficient as a data scientist
As a data scientist, you should spend most of your time working on gaining insights from data, not waiting for your code to finish running. Writing efficient Python code can help reduce runtime and save computational resources, ultimately freeing you up to do the things that have more impact.
In the previous article of this series, “Defining & Measuring Code Efficiency,” I discussed what efficient Python code is and how to use different Python’s built-in data structures, functions, and modules to write cleaner, faster, and more efficient code. I also explored how to time and profile code in order to find bottlenecks.
In this article, we will practice eliminating these bottlenecks and other bad design patterns, using Python’s most used libraries by data scientists: NumPy and pandas.

Table of contents:
Making Your Code Efficient
1.1. Efficiently Combining, Counting, and Iterating
1.2. Introduction to Set Theory
1.3. Eliminating Loops
1.4. Writing Better LoopsIntroduction to Pandas Optimization
2.1. Pandas DataFrame Iteration
2.2. Pandas Alternative to Looping
2.3. Optimal Pandas IteratingReferences
All the codes and datasets used in this article can be found in this GitHub repository
Get All My Books, One Button Away With 40% Off
I have created a bundle for my books and roadmaps, so you can buy everything with just one button and for 40% less than the original price. The bundle features 8 eBooks, including:
1. Making Your Code Efficient
In this section, we will cover more efficiency tips and tricks. You will learn a few useful built-in modules for writing efficient code and practice using set theory. You’ll then learn about looping patterns in Python and how to make them more efficient.
1.1. Efficiently Combining, Counting, and Iterating
Combining Objects
In this subsection, we’ll cover combining, counting, and iterating over objects efficiently in Python. Suppose we have two lists: one of the names and the other is the ages for each of them. We want to combine these lists so that each name is stored next to its age. We can iterate over the names list using enumerate and grab each name's corresponding age using the index variable.
# combining objects
names = ['Ahmed', 'Youssef', 'Mohammed']
age = [25, 27, 40]
combined = []
for i,name in enumerate(names):
combined.append((name, age[i]))
print(combined)But Python’s built-in function zip provides a more elegant solution. The name “zip” describes how this function combines objects like a zipper on a jacket (making two separate things become one). zip returns a zip object that must be unpacked into a list and printed to see the contents. Each item is a tuple of elements from the original lists.
# Combining objects with zip
combined_zip = zip(names, age)
print(type(combined_zip))Python also comes with a number of efficient built-in modules. The collections module contains specialized datatypes that can be used as alternatives to standard dictionaries, lists, sets, and tuples. A few notable specialized datatypes are:
namedtuple: tuple subclasses with named fields
deque: list-like container with fast appends and pops
Counter: dict for counting hashable objects
OrderedDict: dict that retains the order of entries
defaultdict: dict that calls a factory function to supply missing values
Counting Objects
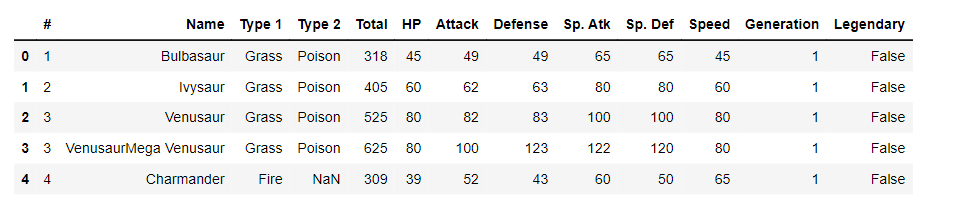
Let’s dig deeper into the Counter object. First, we will upload the Pokémon dataset and print the first five rows of it, then we will count the number of Pokémon per type using a for loop and then using the Counter function.
Let's load the Pokémon dataset and print the first five rows:
import pandas as pd
pokemon = pd.read_csv('pokemon.csv')
pokemon.head()
Now we will count the number of Pokémon per type using loops and calculate the execution time:
%%timeit -r1 -n1
# Counting with for loop
poke_types = pokemon['Type 1']
type_counts = {}
for poke_type in poke_types:
if poke_type not in type_counts:
type_counts[poke_type] = 1
else:
type_counts[poke_type] += 1
print(type_counts)Finally, we will count the number of Pokémon per type using the Counter function and compare the time:
%%timeit -r1 -n1
# counting with collections.Counter()
from collections import Counter
type_counts = Counter(poke_types)
print(type_counts)We can see that using the Counter function from the collections module is a more efficient approach. Just import Counter and provide the object to be counted. No need for a loop!
Counter returns a Counter dictionary of key-value pairs. When printed, it’s ordered by highest to lowest counts. If comparing runtime times, we’d see that using Counter takes less time compared to the standard dictionary approach!
itertools
Another built-in module, itertools, contains functional tools for working with iterators. A subset of these tools is:
Infinite iterators: count, cycle, repeat
Finite iterators: accumulate, chain, zip_longest, etc.
Combination generators: product, permutations, combinations
We will focus on combinatoric generators. These generators efficiently yield Cartesian products, permutations, and combinations of objects. Let’s explore an example.
Combinations
Suppose we want to gather all combinations of Pokémon types. We can do this with a nested for loop that iterates over the poke_types list twice. Notice that a conditional statement is used to skip pairs having the same type twice.
For example, if x is ‘Bug’ and y is ‘Bug’, we want to skip this pair. Since we’re interested in combinations (where order doesn’t matter), another statement is used to ensure that either order of the pair doesn’t already exist within the combos list before appending it. For example, the pair (‘Bug’, ‘Fire’) is the same as the pair (‘Fire’, ‘Bug’). We want one of these pairs, not both.
#Combinations with loop
poke_types = ['Bug', 'Fire', 'Ghost', 'Grass', 'Water']
combos = []
for x in poke_types:
for y in poke_types:
if x == y:
continue
if ((x,y) not in combos) & ((y,x) not in combos):
combos.append((x,y))
print(combos)The combinations generator from itertools provides a more efficient solution. First, we import combinations and then create a combinations object by providing the poke_types list and the length of combinations we desire. Combinations return a combinations object, which we unpack into a list and print to see the result. If comparing runtimes, we’d see that using combinations is significantly faster than the nested loop.
# combinations with itertools
poke_types = ['Bug', 'Fire', 'Ghost', 'Grass', 'Water']
from itertools import combinations
combos_obj = combinations(poke_types, 2)
print(type(combos_obj))
combos = [*combos_obj]
print(combos)1.2. Introduction to Set Theory
Often, we’d like to compare two objects to observe similarities and differences between their contents. When doing this type of comparison, it’s best to leverage a branch of mathematics called set theory. As you know, Python comes with a built-in set data type. Sets come with some handy methods we can use for comparing, such as:
intersection(): all elements that are in both sets
difference(): all elements in one set but not the other
symmetric_difference(): all elements in exactly one set
union(): all elements that are in either set
When we’d like to compare objects multiple times and in different ways, we should consider storing our data in sets to use these efficient methods. Another nice feature of Python sets is their ability to quickly check if a value exists within their members.
We call this membership testing using the in operator. We will go through how using the in operator with a set is much faster than using it with a list or tuple.
Suppose we had two lists of Pokémon names: list_a and list_b, and we would like to compare these lists to see which Pokémon appear in both lists.
We could first use a nested for loop to compare each item in list_a to each item in list_b and collect only those items that appear in both lists. But, iterating over each item in both lists is extremely inefficient.
# Comparing objects with loops
list_a = ['Bulbasaur', 'Charmander', 'Squirtle']
list_b = ['Caterpie', 'Pidgey', 'Squirtle']
in_common = []
for pokemon_a in list_a:
for pokemon_b in list_b:
if pokemon_a == pokemon_b:
in_common.append(pokemon_a)
print(in_common)However, a better way is to use Python’s set data type to compare these lists. By converting each list into a set, we can use the dot-intersection method to collect the Pokémon shared between the two sets. One simple line of code and no need for a loop!
# compaing objects using intersection()
list_a = ['Bulbasaur', 'Charmander', 'Squirtle']
list_b = ['Caterpie', 'Pidgey', 'Squirtle']
set_a = set(list_a)
set_b = set(list_b)
set_a.intersection(set_b)We got the same answer with a much smaller number of code lines. We can also compare the runtime to see how much faster using sets is much faster than using loops.
%%timeit
in_common = []
for pokemon_a in list_a:
for pokemon_b in list_b:
if pokemon_a == pokemon_b:
in_common.append(pokemon_a)
%timeit in_common = set_a.intersection(set_b)We can see that using sets is much faster than using for loops. We can also use a set method to see Pokémon that exist in one set but not in another. To gather Pokémon that exist in set_a but not in set_b, use set_a.difference(set_b).
set_a = {'Bulbasaur', 'Charmander', 'Squirtle'}
set_b = {'Caterpie', 'Pidgey', 'Squirtle'}
set_a.difference(set_b)If we want the Pokémon in set_b, but not in set_a, we use set_b.difference(set_a).
set_a = {'Bulbasaur', 'Charmander', 'Squirtle'}
set_b = {'Caterpie', 'Pidgey', 'Squirtle'}
set_b.difference(set_a)To collect Pokémon that exist in exactly one of the sets (but not both), we can use a method called the symmetric difference.
set_a = {'Bulbasaur', 'Charmander', 'Squirtle'}
set_b = {'Caterpie', 'Pidgey', 'Squirtle'}
set_a.symmetric_difference(set_b)Finally, we can combine these sets using the .union method. This collects all of the unique Pokémon that appear in either or both sets.
set_a = {'Bulbasaur', 'Charmander', 'Squirtle'}
set_b = {'Caterpie', 'Pidgey', 'Squirtle'}
set_a.union(set_b)Another nice efficiency gain when using sets is the ability to quickly check if a specific item is a member of a set’s elements. Consider our collection of 720 Pokémon names stored as a list, a tuple, and a set.
names_list = list(pokemon['Name'])
names_set = set(pokemon['Name'])
names_tuple = tuple(pokemon['Name'])We want to check whether or not the character, Zubat, is in each of these data structures and print the execution time for each data type:
%timeit 'Zubat' in names_list
%timeit 'Zubat' in names_tuple
%timeit 'Zubat' in names_setWhen comparing runtimes, it’s clear that membership testing with a set is significantly faster than with a list or a tuple.
One final efficiency gain when using sets comes from the definition of the set itself. A set is defined as a collection of distinct elements. Thus, we can use a set to collect unique items from an existing object.
Let’s define a primary_types list, which contains the primary types of each Pokémon. If we wanted to collect the unique Pokémon types within this list, we could write a for loop to iterate over the list, and only append the Pokémon types that haven’t already been added to the unique_types list.
primary_types = list(pokemon['Type 1'])
unique_types = []
for prim_type in primary_types:
if prim_type not in unique_types:
unique_types.append(prim_type)
print(unique_types)Using a set makes this much easier. All we have to do is convert the primary_types list into a set, and we have our solution: a set of distinct Pokémon types.
unique_types_set = set(primary_types)
print(unique_types_set)1.3. Eliminating Loops
Although using loops when writing Python code isn’t necessarily a bad design pattern, using extraneous loops can be inefficient and costly. Let’s explore some tools that can help us eliminate the need to use loops in our code. Python comes with a few looping patterns that can be used when we want to iterate over an object’s contents:
For loops iterate over elements of a sequence piece by piece.
While loops execute a loop repeatedly as long as some Boolean condition is met.
Nested loops use multiple loops inside one another.
Although all of these looping patterns are supported by Python, we should be careful when using them. Because most loops are evaluated in a piece-by-piece manner, they are often inefficient solutions.
We should avoid looping as much as possible when writing efficient code. Eliminating loops typically results in fewer lines of code that are easier to understand and interpret. One of the idioms of Pythonic code is that “flat is better than nested.” Striving to eliminate loops in our code will help us follow this idiom.
Suppose we have a list of lists, called poke_stats, that contains statistical values for each Pokémon. Each row corresponds to a Pokémon, and each column corresponds to a Pokémon’s specific statistical value.
Here, the columns represent a Pokémon’s Health Points, Attack, Defense, and Speed. We want to do a simple sum of each of these rows to collect the total stats for each Pokémon.
If we were to use a loop to calculate row sums, we would have to iterate over each row and append the row’s sum to the totals list. We can accomplish the same task in fewer lines of code, with a list comprehension. Or, we could use the built-in map function that we’ve discussed previously in the previous article.
# List of HP, Attack, Defense, Speed
poke_stats = [
[90, 92, 75, 60],
[25, 20, 15, 90],
[65, 130, 60, 75],
]
%%timeit
totals = []
for row in poke_stats:
totals.append(sum(row))%timeit totals_comp = [sum(row) for row in poke_stats]%timeit totals_map = [*map(sum, poke_stats)]Each of these approaches will return the same list, but using a list comprehension or the map function takes one line of code and has a faster runtime.
We’ve also covered a few built-in modules that can help us eliminate loops in the previous article. Instead of using the nested for loop, we can use combinations from the itertools module for a cleaner, more efficient solution.
poke_types = ['Bug', 'Fire', 'Ghost', 'Grass', 'Water']
# Nested for loop approach
combos = []
for x in poke_types:
for y in poke_types:
if x == y:
continue
if ((x,y) not in combos) & ((y,x) not in combos):
combos.append((x,y))
# Built-in module approach
from itertools import combinations
combos2 = [*combinations(poke_types, 2)]Another powerful technique for eliminating loops is to use the NumPy package. Suppose we had the same collection of statistics we used in a previous example, but stored in a NumPy array instead of a list of lists.
We’d like to collect the average stat value for each Pokémon (or row) in our array. We could use a loop to iterate over the array and collect the row averages.
%%timeit
avgs = []
for row in poke_stats:
avg = np.mean(row)
avgs.append(avg)Eliminate Loops with NumPy
But NumPy arrays allow us to perform calculations on entire arrays all at once. Here, we use the dot-mean method and specify an axis equal to 1 to calculate the mean for each row (meaning we calculate an average across the column values). This eliminates the need for a loop and is much more efficient.
%timeit avgs = poke_stats.mean(axis=1)When comparing runtimes, we see that using the dot-mean method on the entire array and specifying an axis is significantly faster than using a loop.
1.4. Writing Better Loops
We’ve discussed how loops can be costly and inefficient. But sometimes you can’t eliminate a loop. In this section, we’ll explore how to make loops more efficient when looping is unavoidable.
Before diving in, some of the loops we’ll discuss can be eliminated using techniques covered in previous lessons. For demonstrative purposes, we’ll assume the use cases shown here are instances where a loop is unavoidable.
The best way to make a loop more efficient is to analyze what’s being done within the loop. We want to make sure that we aren’t doing unnecessary work in each iteration.
If a calculation is performed for each iteration of a loop, but its value doesn’t change with each iteration, it’s best to move this calculation outside (or above) the loop.
If a loop is converting data types with each iteration, it’s possible that this conversion can be done outside (or below) the loop using a map function. Anything that can be done once should be moved outside of a loop. Let’s explore a few examples.
Moving calculations above a loop
We have a list of Pokémon names and an array of each Pokémon’s corresponding attack value. We’d like to print the names of each Pokémon with an attack value greater than the average of all attack values.
To do this, we’ll use a loop that iterates over each Pokémon and its attack value. For each iteration, the total attack average is calculated by finding the mean value of all attacks. Then, each Pokémon’s attack value is evaluated to see if it exceeds the total average.
import numpy as np
names = ['Absol', 'Aron', 'Jynx', 'Natu', 'Onix']
attacks = np.array([130, 70, 50, 50, 45])
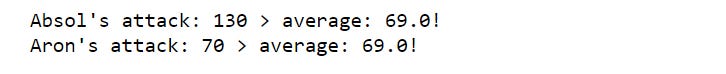
for pokemon,attack in zip(names, attacks):
total_attack_avg = attacks.mean()
if attack > total_attack_avg:
print(
"{}'s attack: {} > average: {}!"
.format(pokemon, attack, total_attack_avg)
)The inefficiency in this loop is the total_attack_avg variable being created with each iteration of the loop. But this calculation doesn’t change between iterations since it is an overall average.
We only need to calculate this value once. By moving this calculation outside (or above) the loop, we calculate the total attack average only once. We get the same output, but this is a more efficient approach.
import numpy as np
names = ['Absol', 'Aron', 'Jynx', 'Natu', 'Onix']
attacks = np.array([130, 70, 50, 50, 45])
# Calculate total average once (outside the loop)
total_attack_avg = attacks.mean()
for pokemon,attack in zip(names, attacks):
if attack > total_attack_avg:
print(
"{}'s attack: {} > average: {}!"
.format(pokemon, attack, total_attack_avg)
)%%timeit
for pokemon,attack in zip(names, attacks):
total_attack_avg = attacks.mean()
if attack > total_attack_avg:
print(
"{}'s attack: {} > average: {}!"
.format(pokemon, attack, total_attack_avg)
)
%%timeit
# Calculate total average once (outside the loop)
total_attack_avg = attacks.mean()
for pokemon,attack in zip(names, attacks):
if attack > total_attack_avg:
print(
"{}'s attack: {} > average: {}!"
.format(pokemon, attack, total_attack_avg)
)We see that keeping the total_attack_avg calculation within the loop takes more than 120 microseconds.
Holistic conversions
Another way to make loops more efficient is to use holistic conversions outside (or below) the loop. In the example below, we have three lists from the 720 Pokémon dataset: a list of each Pokémon’s name, a list corresponding to whether or not a Pokémon has a legendary status, and a list of each Pokémon’s generation.
We want to combine these objects so that each name, status, and generation is stored in an individual list. To do this, we’ll use a loop that iterates over the output of the zip function.
Remember, zip returns a collection of tuples, so we need to convert each tuple into a list since we want to create a list of lists as our output. Then, we append each poke_list to our poke_data output variable. By printing the result, we see our desired list of lists.
%%timeit
poke_data = []
for poke_tuple in zip(names_list, legend_status_list, generations_list):
poke_list = list(poke_tuple)
poke_data.append(poke_list)However, converting each tuple to a list within the loop is not very efficient. Instead, we should collect all of our poke_tuples together and use the map function to convert each tuple to a list.
The loop no longer converts tuples to lists with each iteration. Instead, we moved this tuple to list conversion outside (or below) the loop. That way, we convert data types all at once (or holistically) rather than converting in each iteration.
%%timeit
poke_data_tuples = []
for poke_tuple in zip(names_list, legend_status_list, generations_list):
poke_data_tuples.append(poke_tuple)
poke_data = [*map(list, poke_data_tuples)]Runtimes show that converting each tuple to a list outside of the loop is more efficient.
2. Introduction to Pandas Optimization
2.1. Pandas DataFrame Iteration
Pandas is a library used for data analysis. The main construct of pandas is the DataFrame, a tabular data structure with labeled rows and columns. We will focus on the best approaches for iterating over a DataFrame.
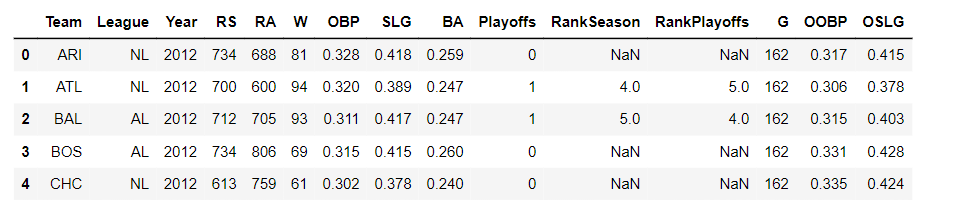
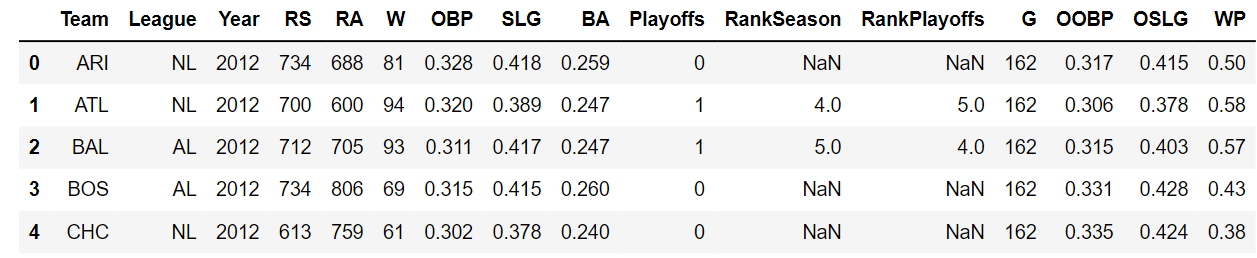
Let’s begin by analyzing a Major League Baseball dataset. It contains the team stats for each Major League Baseball team from the year 1962 to 2012, which are stored in a pandas DataFrame named baseball_df.
import pandas as pd
baseball_df = pd.read_csv('baseball_stats.csv')
baseball_df.head()We will focus on the W column, which specifies the number of wins a team had in a season, and the G column, which contains the number of games a team played in a season.
Calculating & Adding win percentage
One popular statistic used to evaluate a team’s performance for a given season is the team’s win percentage. This metric is calculated by dividing a team’s total wins by the number of games played. Here is a simple function to perform this calculation.
def calc_win_perc(wins, games_played):
win_perc = wins / games_played
return np.round(win_perc,2)Now we would like to create a new column in our baseball_df DataFrame that stores each team’s win percentage for a season. To do this, we’ll need to iterate over the DataFrame’s rows and apply our calc_win_perc function. First, we create an empty win_perc_list to store all the win percentages we’ll calculate.
Then, we write a loop that will iterate over each row of the DataFrame. Notice that we are using an index variable (i) that ranges from zero to the number of rows that exist within the DataFrame. We then use the .iloc method to look up each individual row within the DataFrame using the index variable.
Then we grab each team’s wins and games played by referencing the W and G columns. Next, we pass the team’s wins and games played to calc_win_perc to calculate the win percentages.
Finally, we append win_perc to win_perc_list and continue the loop. We create our desired column in the DataFrame, called WP, by setting the column value equal to the win_perc_list.
%%timeit
win_perc_list = []
for i in range(len(baseball_df)):
row = baseball_df.iloc[i]
wins = row['W']
games_played = row['G']
win_perc = calc_win_perc(wins, games_played)
win_perc_list.append(win_perc)
baseball_df['WP'] = win_perc_list
baseball_df.head()Looping over the DataFrame with .iloc gave us our desired output, but is it inefficient? When estimating the runtime, the .iloc approach took 660 milliseconds, which is pretty inefficient.
Iterating with .iterrows()
Pandas come with a few efficient methods for looping over a DataFrame. The first method we’ll cover is the .iterrows method. This is similar to the .iloc method, but .iterrows returns each DataFrame row as a tuple of (index, pandas Series) pairs.
This means each object returned from .iterrows contains the index of each row as the first element and the data in each row as a pandas Series as the second element.
Notice that we still create the empty win_perc_list, but now we don’t have to create an index variable to look up each row within the DataFrame. .iterrows handles the indexing for us! The remainder of the for loop stays the same to create a new win percentage column within our baseball_df DataFrame.
%%timeit
win_perc_list = []
for i,row in baseball_df.iterrows():
wins = row['W']
games_played = row['G']
win_perc = calc_win_perc(wins, games_played)
win_perc_list.append(win_perc)
baseball_df['WP'] = win_perc_listUsing .iterrows takes roughly half the time .iloc takes to iterate over our DataFrame.
Iterating with .itertuples()
Pandas also come with another iteration method called .itertuples, which is often more efficient than .iterrows. Let’s continue using our baseball dataset to compare these two methods. Suppose we have a pandas DataFrame called team_wins_df that contains each team’s total wins in a season.
If we use .iterrows to loop over our team_wins_df DataFrame and print each row’s tuple, we see that each row’s values are stored as a pandas Series. Remember, .iterrows returns each DataFrame row as a tuple of (index, pandas Series) pairs, so we have to access the row’s values with square bracket indexing.
But, we could use .itertuples to loop over our DataFrame rows instead. The .itertuples method returns each DataFrame row as a special data type called a namedtuple.
A namedtuple is one of the specialized data types that exist within the collections module we’ve discussed previously. These data types behave just like a Python tuple but have fields accessible using attribute lookup.
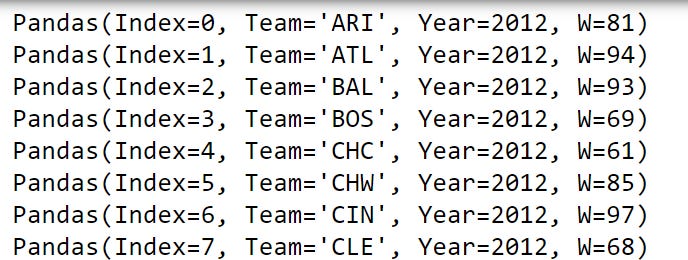
team_wins_df = baseball_df[['Team','Year','W']]
for row_namedtuple in team_wins_df.itertuples():
print(row_namedtuple)Notice in the output that each printed row_namedtuple has an Index attribute, and each column in our team_wins_df has an attribute. That means we can access each of these attributes with a lookup using a dot method.
Here, we can print the last row_namedtuple’s Index using row_namedtuple.Index. We can print this row_namedtuple’s Team with row_namedtuple.Team, Year with row_namedtuple.Year and so on.
print(row_namedtuple.Index)print(row_namedtuple.Team)When we compare .iterrows to .itertuples, we see that there is quite a bit of improvement!
%%timeit
for row_tuple in team_wins_df.iterrows():
print(row_tuple)%%timeit
for row_namedtuple in team_wins_df.itertuples():
print(row_namedtuple)The reason .itertuples is more efficient than .iterrows is due to the way each method stores its output. Since .iterrows returns each row’s values as a pandas Series, there is a bit more overhead.
2.2. Pandas Alternative to Looping
In order to write efficient code, we want to avoid looping as much as possible. Therefore, now we will explore an alternative to using .iterrows and .itertuples to perform calculations on a DataFrame.
We’ll continue using the baseball dataset. We will first create the calc_run_diff function. This function calculates a team’s run differential for a given year by subtracting the team’s total number of runs allowed from its total number of runs scored in a season.
def calc_run_diff(runs_scored, runs_allowed):
run_diff = runs_scored - runs_allowed
return run_diffWe’d like to create a new column in our baseball_df DataFrame called RD that stores each team’s run differentials over the years. Previously, we did this with a for loop using either .iterrows or .itertuples. Here, we’ll use .iterrows as an example.
Notice that we are iterating over baseball_df with a for loop, passing each row’s RS and RA columns into our calc_run_diff function, and then appending each row’s result to our run_diffs_iterrows list. This gives us our desired output, but it is not our most efficient option.
Pandas.apply() Method
One alternative to using a loop to iterate over a DataFrame is to use pandas.apply() method. This function acts like the map function. It takes a function as an input and applies this function to an entire DataFrame. Since we are working with tabular data, we must specify an axis that we would like our function to act on.
Using a zero for the axis argument will apply our function on columns, while using a one for the axis argument will apply our function on all rows. Just like the map function, pandas.apply() method can be used with anonymous functions or lambdas.
Let’s walk through how we’d use the .apply() method to calculate our run differentials. First, we call .apply() on the baseball_df DataFrame. Then, we use a lambda function to iterate over the rows of the DataFrame.
Notice that our argument for lambda is row (since we are applying to each row of the DataFrame). For every row, we grab the RS and RA columns and pass them to our calc_run_diff function. Lastly, we specify our axis to tell dot-apply that we want to iterate over rows instead of columns.
%%timeit
run_diffs_apply = baseball_df.apply(
lambda row: calc_run_diff(row['RS'], row['RA']),
axis=1)
baseball_df['RD'] = run_diffs_applyWhen we use the .apply method to calculate our run differentials, we don’t need to use a for loop. We can collect our run differentials directly into an object called run_diffs_apply.
We can see that the .iterrows approach took about 321 milliseconds to complete. But, using the dot-apply method took only 45.2 milliseconds. A definite improvement!
2.3. Optimal Pandas Iterating
We’ve come a long way from our first .iloc approach for iterating over a DataFrame. Each approach we’ve discussed has really improved the performance.
But these approaches focus on performing calculations for each row of our DataFrame individually. Now we will explore some Panda's internals that allow us to perform calculations more efficiently.
Since pandas is a library that is built on NumPy. This means that each pandas DataFrame we use can take advantage of the efficient characteristics of NumPy arrays discussed in the previous article.
We discussed broadcasting and how it allows the NumPy arrays to vectorize operations, so they are performed on all elements of an object at once.
This allows us to efficiently perform calculations over entire arrays. Just like NumPy, pandas are designed to vectorize calculations so that they operate on entire datasets at once (not just on a row-by-row basis). Let’s explore this concept with some examples.
We’ll continue to use the baseball_df DataFrame we have been using throughout the chapter. Since pandas are built on top of NumPy, we can grab any of these DataFrame columns’ values as a NumPy array using the dot-values method.
Here, we are collecting the W column’s values into a NumPy array called wins_np. When we print the type of wins_np, we see that it is, in fact, a NumPy array.
wins_np = baseball_df['W'].values
print(type(wins_np))We can see the contents of the array by printing it and verifying that it is the same as the W column from our DataFrame.
print(wins_np)The beauty of knowing that pandas are built on NumPy can be seen when taking advantage of a NumPy array’s broadcasting abilities. Remember, this means we can vectorize our calculations and perform them on entire arrays all at once! Instead of looping over a DataFrame and treating each row independently, like we’ve done with .iterrows, .itertuples, and .apply, we can perform calculations on the underlying NumPy arrays of our baseball_df DataFrame.
Here, we gather the RS and RA columns in our DataFrame as NumPy arrays, and use broadcasting to calculate run differentials all at once!
%%timeit
run_diffs_np = baseball_df['RS'].values - baseball_df['RA'].values
baseball_df['RD'] = run_diffs_npWhen we use NumPy arrays to perform our run differential calculations, we can see that our code becomes much more readable. Here, we can explicitly see how our run differentials are being calculated. Also, when we time our NumPy arrays approach, we see that our run differential calculations take microseconds!
All other approaches were reported in milliseconds. Our array approach is orders of magnitude faster than all previous approaches! Using a DataFrame’s underlying NumPy arrays to perform calculations can help us gain some massive efficiencies.
This newsletter is a personal passion project, and your support helps keep it alive. If you would like to contribute, there are a few great ways:
Subscribe. A paid subscription helps to make my writing sustainable and gives you access to additional content.*
Grab a copy of my book Bundle. Get my 7 hands-on books and roadmaps for only 40% of the price
Thanks for reading, and for helping support independent writing and research!
Are you looking to start a career in data science and AI, but do not know how? I offer data science mentoring sessions and long-term career mentoring:
Mentoring sessions: https://lnkd.in/dXeg3KPW
Long-term mentoring: https://lnkd.in/dtdUYBrM



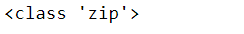



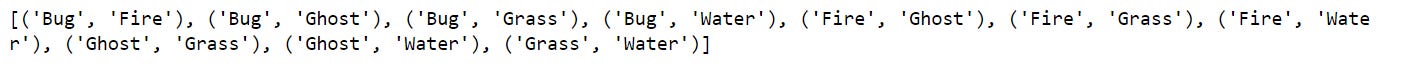


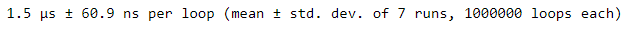
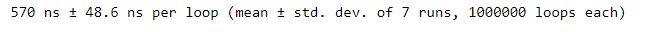


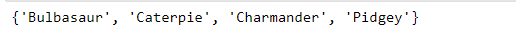


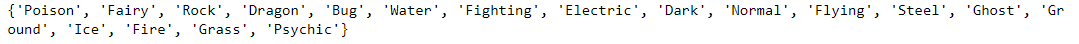
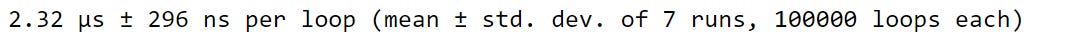



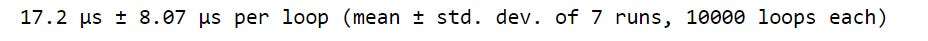


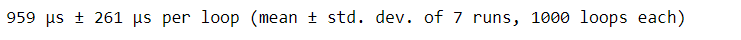


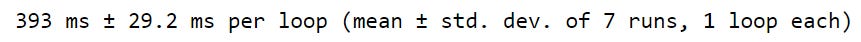





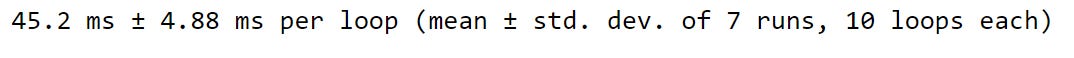
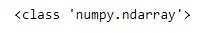



Something that helped me when I was getting started: start thinking of DataFrames not as spreadsheets to loop through, but as physical material you're shaping. .apply() is like using a precision tool on each row, but vectorization is like putting the whole thing through a machine press. It's a different kind of operation entirely.
The set operations section is a hidden gem. I've seen a lot of junior analysts write nested loops for data validation that could be a one-liner with set.difference(). It's one of the easiest wins for both performance and code clarity.